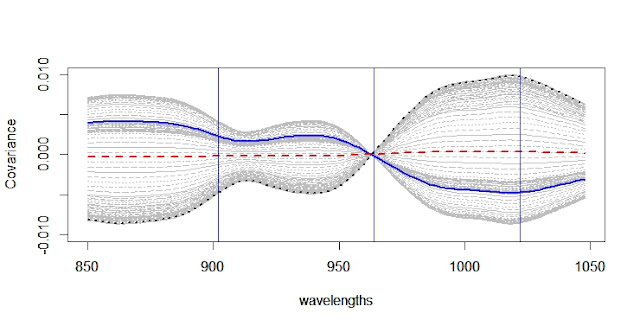
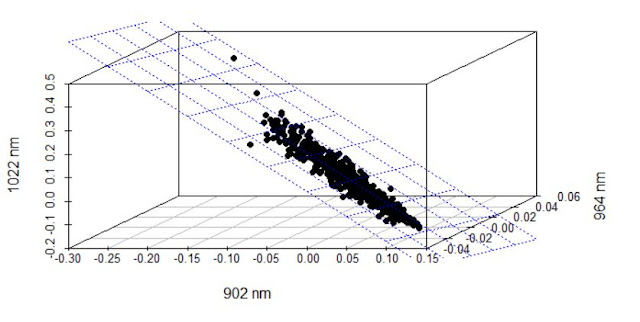
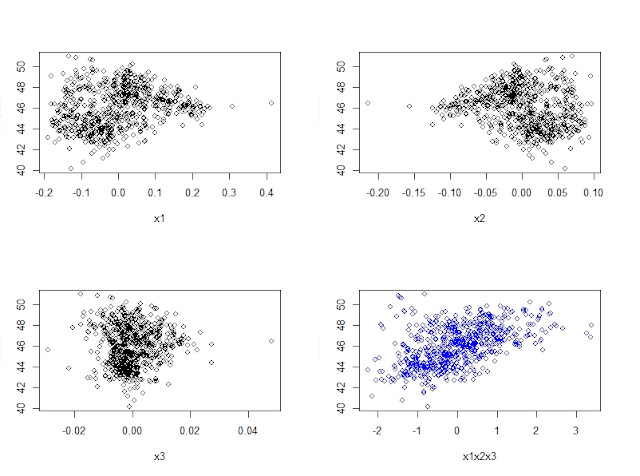
When developing a LOCAL calibration in Win ISI, we use an input file and a Library file with the idea the idea to select the best possible model, so the selection of a well representative input (let´s call validation set) is very important to have success in the development of the model.
So the model is conditioned to the input file, if we have choose another input file we could have get another model which performs different, so the need of a Test Set is obvious to check how the model performs with new data.
It is important to have this in mind, so one proposal would be to divide (randomly) the data in three sets: 60% for training, 20% for Input or Validation, and another 20% for testing.
There are other ways to sort the data in order to select these three Sets (time, seasons, species,...). One thing is clear, some of the models developed will perform better than others, so you can keep several of them and you can check this when you have new data and use an statistic an MSE (Mean Square Error) to compare them.