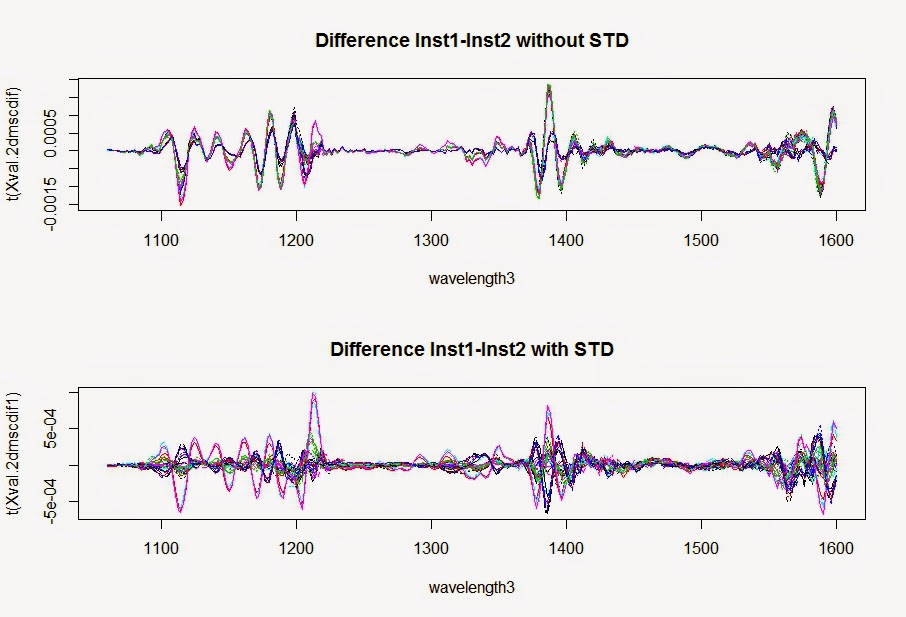
All the samples in ISI Scan are stored in the "data" folder in the ISI Scan directory. Keeping the Access data base, and files included into that data folder is important to run the ISI Scan fast and secure.
So it is important to make backups regularly and clean the database at the same time. All this is configures at the System Profile window.
In this window, we configure the samples the samples which stay in the database and the samples which are exported or deleted definitively.
Spectra of samples with Lab Value attached are always kept in ISI Scan , so they can be exported as CAL files or used for Monitor purposes.
Read the ISI Scan Help section of System profile for a better understanding of the procedure.
After the Backup, a window appears searching for samples to archive:
Samples are archived in a new folder called Archive inside the Backup folder. Into this Archive folder there are new folders with the name of the products and inside this folders are the NIR files with the spectra, and the ANL or CSV files with the results. So you can export this files to Win ISI to work with them.
After the Backup, a window appears searching for samples to archive:
Samples are archived in a new folder called Archive inside the Backup folder. Into this Archive folder there are new folders with the name of the products and inside this folders are the NIR files with the spectra, and the ANL or CSV files with the results. So you can export this files to Win ISI to work with them.
If the samples stay in the Data folder of ISI Scan and are not yet archived, we can search them clicking with the mouse on Products and selecting "Search". A new window appears where we have filter options, once searched the samples will go to the "Selected Samples" folder.