I have spectra from sunflower seed grinded from 3 NIR instruments (range 400-2500 nm). I prepare the data frame separating the spectral range in two segments (Segment 1 or VIS from 400-1100 nm, and segment 2 or NIR from 1100 to 2500). Reference values are from four different laboratories.
In a previous post I have transformed the NIR raw spectra to MSC, using a function from the R package "Chemometrics with R".
In this post I want to run a regression with PLS (PLSR) using the Segment 2, and with the math treatment MSC.
sflw1 <- plsr(G00rmn~NIRmsc, ncomp = 10,data =sflw.msc2 ,
validation = "LOO")
sflw1 <- plsr(G00rmn~NIRmsc, ncomp = 10,data =sflw.msc2 ,
validation = "LOO")
VALIDATION: RMSEP
Cross-validated using 107 leave-one-out segments.
(Intercept) 1 comps 2 comps 3 comps 4 comps 5 comps 6 comps
CV 2.87 2.465 2.074 1.112 1.038 0.9903 0.9833
adjCV 2.87 2.465 2.074 1.111 1.038 0.9899 0.9829
7 comps 8 comps 9 comps 10 comps
CV 0.9746 0.9781 0.9754 0.9683
adjCV 0.9741 0.9775 0.9745 0.9675
TRAINING: % variance explained
1 comps 2 comps 3 comps 4 comps 5 comps 6 comps 7 comps 8 comps
X 41.93 89.86 95.78 97.02 98.21 99.21 99.62 99.76
G00rmn 31.47 51.80 86.47 89.14 90.36 90.64 91.01 91.42
9 comps 10 comps
X 99.81 99.89
G00rmn 92.30 92.62
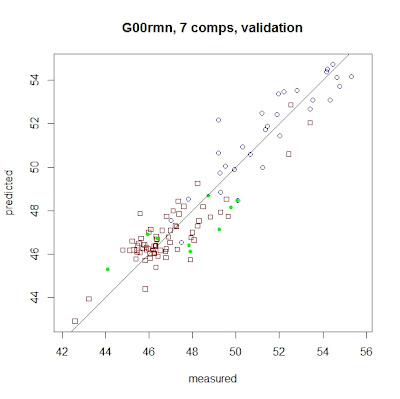
And
now we can see the X-Y plot for the LOO Regression (with 7 comps), with
different colors and symbols for the samples from the different instruments.
plot(sflw1, ncomp = 7, asp = 1, line = TRUE,
pch=c(20:22)[sflw.msc2$Operator],
col=c("green","blue","brown")[sflw.msc2$Operator])
col=c("green","blue","brown")[sflw.msc2$Operator])

No hay comentarios:
Publicar un comentario