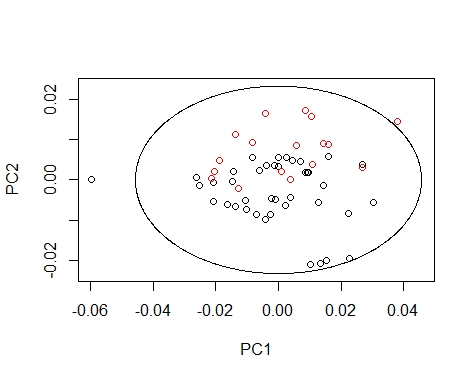
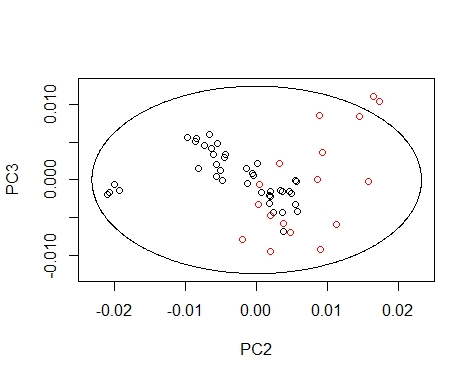
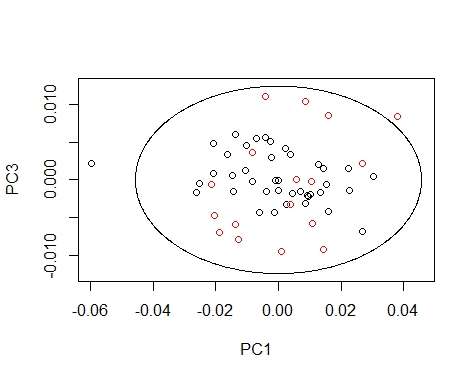
We can coloured the scores by their season value, so we can see how every season extend the scores in the different principal components maps.
fish_1d_all_pc<-nipals(fish_1d_all$nir_1d,10)
rownames(fish_1d_all_pc$T)<-fish_1d_all$Sample
colnames(fish_1d_all_pc$T)<-seq(1,10,by=1)
##### Creating a list for NIPALS
library(chemometrics)
fish12_nipals <-list(scores=fish_1d_all_pc$T,
loadings=fish_1d_all_pc$P,
sdev=apply(fish_1d_all_pc$T, 2, sd))
Now we plot the orthogonal and scores distances and we check which ones are outside certain limits, coulouring the samples by their season and using 5 principal components.
res<-pcaDiagplot(fish_1d_all$nir_1d,X.pca=fish12_nipals,
a=5,col=fish_1d_all$Season)
Samples outside the cutoff are the Mahalanobis distance outliers considering 5 principal components.
We can draw the Mahalanobis Ellipses with the combinations: PC1 vs PC2,
PC1 vs PC3, and PC2 vs PC3. Samples are coloured by their season (black for fish1 and red for fish2).
library(chemometrics)
par(mfrow=c(1,1))####### PC2 vs PC3 ##########
drawMahal(fish_1d_all_pc$T[,c(1,2)],
center=apply(fish_1d_all_pc$T[,c(1,2)],2,mean),
covariance=cov(fish_1d_all_pc$T[,c(1,2)]),
quantile=0.975,xlab="PC1",ylab="PC2",
####### PC2 vs PC3 ##########
drawMahal(fish_1d_all_pc$T[,c(2,3)],
center=apply(fish_1d_all_pc$T[,c(2,3)],2,mean),
covariance=cov(fish_1d_all_pc$T[,c(2,3)]),
quantile=0.975,xlab="PC2",ylab="PC3",
col=fish_1d_all$Season)
####### PC1 vs PC3 ########
drawMahal(fish_1d_all_pc$T[,c(1,3)],
center=apply(fish_1d_all_pc$T[,c(1,3)],2,mean),
covariance=cov(fish_1d_all_pc$T[,c(1,3)]),
quantile=0.975,xlab="PC1",ylab="PC3",
col=fish_1d_all$Season)
Samples out of the ellipse can be identified with the identify function but that does not mean that it is an outlier because that depends of a general computation between all the principal components.
As you can see all the process is becoming very straightforward as we continue with more posts of more fishing seasons.