These
five samples are the same in the Training Set scanned in Instrument 1 and the
Training Set scanned in Instrument 2, so it is clear that the problem is that
the lab value does not correlate as the others with the spectra.
First,
we remove the samples from the Training Set 1:
nir.tr1a.2dmsc<-nir.tr1.2dmsc[c(-19,-122,-126,-127,-150),]
Now, the new regression model without outliers,
and with the math treatments we consider apropiate as MSC + Second derivative:
mod3a<-plsr(Y~X,data=nir.tr1a.2dmsc,ncomp=10,validation="LOO")
Comparing the summaries of the models with
and without outliers we see the logical improvement.
nir.tr2a.2dmsc<-nir.tr2.2dmsc[c(-19,-122,-126,-127,-150),]
tr2a.pred<-as.matrix(predict(mod3a,ncomp=3,newdata=nir.tr2a.2dmsc))monit.tr2a<-cbind(nir.tr2a.2dmsc$Y,tr2a.pred)
monit.tr2 colnames(monit.tr2a)<-c("Y.tr.lab","Y.tr2.pred")
monit.tr2a<-round(monit.tr2a,digits=1)
Now with
this table we can run the Monitor function:
monitor14(monit.tr2a[,2],monit.tr2a[,1],150,3,0.95,2.904)
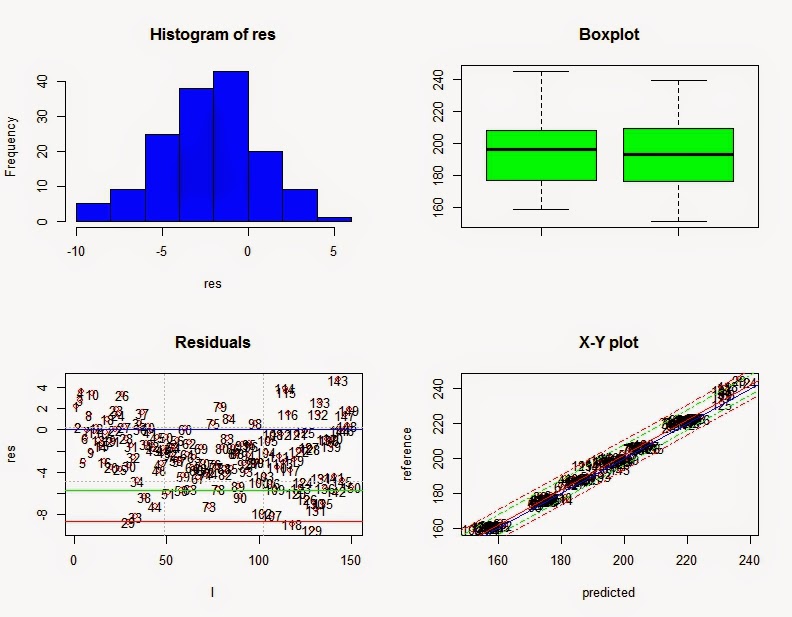
The results show an improvement in the
RMSEP and the SEP statistic tell us the error corrected by the bias. The monitor function now recommend a Bias adjustment.
The distribution of the residuals shows the bias problem, but it is quite uniform once we correct the bias.
-------------------------------------
N Validation Samples = 150
N Calibration Samples = 150
N Calibration Terms = 3
-------------------------------------
RMSEP : 3.642
Bias : -2.249
SEP : 2.875
UECLs : 3.327
***SEP is bellow BCLs (O.K)***
Corr : 0.9917
RSQ : 0.9834
Slope : 1.002
Intercept: 1.874
RER : 29.92 Good
RPD : 7.759 Very Good
BCL(+/-): 0.4637
***Bias adjustment is recommended***
Residual Std Dev is : 2.884
***Slope adjustment in not necessary***

No hay comentarios:
Publicar un comentario