27 ene 2022
Eye on Agriculture Today: Soil Texture By Feel
24 ene 2022
PCA with the first derivative
In a previous post we had calculated the PCA with the
math treatment SNV+Detrend and we calculated a first sample set of outliers
with the Mahalanobis distance.
When calculating PCA we have to treat as best the
spectra as possible in order to detect populations or boundaries and if we
treat the spectra with math treatments which help to do this task is great. So indeed,
to apply the SNV + Detrend, I apply this time the first derivative to the
spectra (soil from Spanish soil from LUCAS database) and calculate the PCA.
We can have a look to the score’s
maps (six PC recommended) to find if there are boundaries on them:
Look at the maps which include
PC2 as one of the axes. There are a certain number of samples which takes a
different direction than the rest. This second PC term can be useful to find
something interesting on the spectra.
To see what is happening we can see the loading spectra for this second PC term:
This loading spectra has the
first derivative math treatment applied, so we can compare it with a library of
reference known spectra (minerals in this case) to see which is the best match,
and in this case the best match is with the gypsum mineral, so this second
term is explaining part of the variance included in our spectra database due to
the addition of gypsum to the soil.
23 ene 2022
LUCAS SP Database vs. pure mineral Gypsum
In order to find if the soil has traces of a certain mineral, it is useful to overplot our soil samples (in this case the soil Spanish samples included in the LUCAS database, with the pure mineral spectrum (in this case Gypsum). We must overplot them with the same math treatment and in the same scale. I do it, in this case, with the spectra treated with the SG second derivative.
Second derivative is quite helpfull to find these matches. If we do it, for example with SNV+Detrend, some of the bands are hiden by other samples and the assumption that there are samples with gypsum is less clear.
13 ene 2022
Detecting outliers with Mahalanobis distance
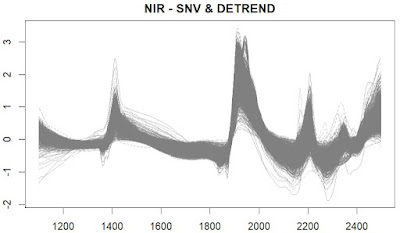
In this first plot we see the spectra of the LUCAS spanish database treated with the SNV and Detrend math treatment of the "Prospectr" package, where we remove the quadratic trend (as we saw in the last post):
11 ene 2022
Quadratic trendlines
In the last post, I talk about the use of the function "detrend" in the package "prospectr" to remove the quadratic trend lines in the soil NIR spectra. We can see these trendlines overplotted to the spectra (I do it just for five of the soil spectra from the Spanish soil LUCAS database).
In red is the spectra treated with SNV and in blue the quadratic trend lines to apply to the SNV treated spectra, to remove them and convert them in a SNV + Detrend spectra.
9 ene 2022
Scale and linear / Scale and quadratic
Scale and linear and Scale and quadratic are two of the scatter corrections
that we can find in Win ISI 4, and they are used to remove as much as possible
the multiplicative scatter we found in the NIR spectra. Two are the packages
that have a detrend function in R, so depending on the case of the multiplicative
effect (linear or quadratic, we can use the pracma or prospectr packages.
The detrend function of the Prospectr package is equivalent to the math
treatment in WinISI 4 called "scale and quadratic” and combine the SNV
treatment with the remove of a quadratic trendline in the spectra. In the case of
the combination of SNV and Detrend from the Pracma package we obtain the Win
ISI math treatment "Scale and Linear" and in this case we remove the
linear trend line due to the scatter. We can try both to check which one gives
the better performance.
I show the spectra of both treatments applied to the soil LUCAS database
spectra filtered for the Spanish soils:
Samples are coloured according the soil type (cropland, grassland,
woodland,....). The jump in the detector change it is common due that we treat
individually the math treatment for the NIR and VIS ranges (trend are different in VIS and NIR regions).
5 ene 2022
Trim spectra from LUCAS database
The spectra from LUCAS database (ESDAC*) seem to come from a XDS (VIS + NIR) instrument, and this instrument give two options when exporting the spectra (every 0.5 nm, and every 2 nm), the data comes in 0.5 nm, and that means that we have 4200 data poits per spectrum, and that means a huge spectra matrix.
We can trim the spectra keeping just the spectral data every two nanometers, so we will have the reflectance values from 400 to 2498 nm every 2nm, so we have a less heavy matrix of 1050 data points per spectrum. This fuction will give us the spectra like a NIR6500.
Just create a sequence to select one column of every four, and call that function (like in my case) "trim05to2":
dim(lucas_spain$spc)2604 4200
spec2nm <- trim05to2(lucas_spain$spc)
dim(spec2nm)
2604 1050
Importing LUCAS database from R into WinISI
write.table(spec2nm, file="lucas_spain.txt",
row.names=TRUE,
col.names=FALSE)
This way we have
the samples IDs, but we don´t need the column name (wavelengths), because Win
ISI will create them when importing the spectra.
Once we have the txt file, we convert it to a ".nir" file which is
the spectra format for Win ISI with the Win ISI tool "Convert",
selecting the configuration from TXT to Win ISI.
After
filling the options Win ISI ask, we wait for the conversion succeeded
message and we have the ".nir" file ready to view: