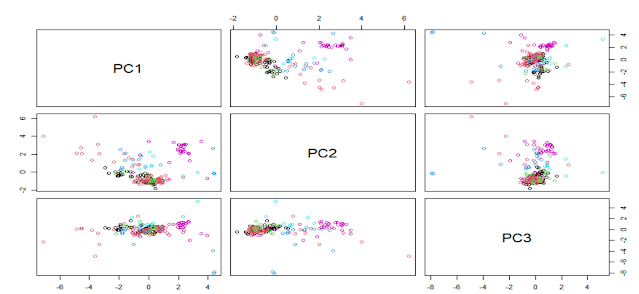
In the previous plot we have seen the score plots trying to see some clusters or looking for some groupings based on the "Type" variable. We have seen the correlation plot as well for the Glass data set and now it is time for the loadings plots.
The loadings plots are useful to understand how the predictors are associated with the several components. In the case of the Glass data set we can see the predictors weights for the first 3 principal components:
As we saw in the correlation plot (in a previous post) RI and CA are very close in both plots and that means that they are highly correlated. We can se how "Ba" and "Mg" are inverse correlated and have a high weight for PC2 and almost not weight for PC3. There are a lot of conclusions we can take out for the loading plots.
In blue the code I use to get the plots:
plot(transformed[,2:3], col = Type, pch = 1,
xlab = paste("PC 1 (", variance[1], "%)", sep = ""),
ylab = paste("PC 2 (", variance[2], "%)", sep = ""))
plot(trans$rotation[,1],trans$rotation[,2],type = "n",
xlab = paste("PC 1 (", variance[1], "%)", sep = ""),
ylab = paste("PC 2 (", variance[2], "%)", sep = ""))
text(trans$rotation[,1:3], labels = rownames(trans$rotation))
plot(transformed[,3:4], col = Type, pch = 1,
xlab = paste("PC 2 (", variance[2], "%)", sep = ""),
ylab = paste("PC 3 (", variance[3], "%)", sep = ""))
plot(trans$rotation[,2],trans$rotation[,3],type = "n",
xlab = paste("PC 2 (", variance[2], "%)", sep = ""),
ylab = paste("PC 3 (", variance[3], "%)", sep = ""))
text(trans$rotation[,2:3], labels = rownames(trans$rotation))