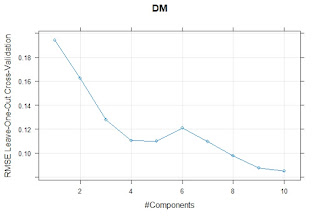
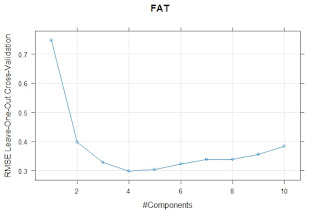
I have a dataset with values for different types of Iberian ham of different qualities analyzed by the reference methods and by a Near Infrared Transmission Method. We are using for the prediction a Meat Calibration Model where probably this type of samples are not represented so some adjustments can be necessary, and we want to check if its better to do it in general or individually by types of ham.
We have data for moisture, protein, fat, salt and water activity. This data is not cleaned, so we will have to remove probably some samples due to a wrong lab values or samples not well presented to the instrument due to several reasons.
We can start by Moisture in this post, and we can plot on the X axis the Predicted Value and on the Y axis the lab Value. We use colors to see in a better way how the samples fit in the plot and from the plot we can take conclusions about which types of products have less or more water.
ggplot(ham_data,aes(x = Moi_FSC, y= Moi_LAB,color = Product)) +
geom_point()
Let´s look to the data by product, so we can have a better knowledge about every product:
ggplot(jamon_data,aes(x = Moi_FSC, y= Moi_LAB,color = Product)) +
geom_point() +
facet_wrap(~Product)
If we have all this information in plots for the different constituents and product we can make a visual understanding of every product. We can check apart the statistics later , but it is clear that the impact of the tydyverse plots is great.