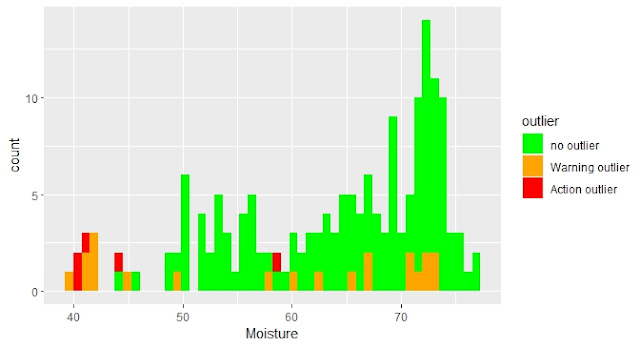
Hemos visto los anómalos de distancia de mahalanobis marcados en los histogramas para los distintos parámetros. Veamos estos mismos anómalos marcados en los gráficos de inter-correlación entre parámetros y en los espectros con su correspondiente tratamiento matemático.
Todas estas observaciones previas al desarrollo de los modelos de calibración nos pueden ser muy útiles.20 dic 2022
PCA - NIT Tutorial (part 12)
16 dic 2022
PCA - NIT Tutorial (part 10)
click the image to access to the Quarto blog post.
En este post trato el tema del cálculo de la distancia de Mahalanobis en el espacio de componentes principales y cómo podemos visualizarla gráficamente con elipses en sus planos o también con cubos, marcando los anómalos con distancias superiores al umbral que determinemos.
In this post I talk about the Mahalanobis distance calculation in the Principal Component Space and how we can look at graphically with ellipses in the PCA scores maps planes or also in 3D with cubes.
PCA - NIT tutorial (part 11)
Does exist a relationship between the Robust Mahalanobis distances and the concentration of the parameters?. Click the image to access to the Quarto blog post which explain it.
3 nov 2022
PCA - NIT Tutorial (Part 9)
In this post I continue exploring the PCA looking to the scores maps and
creating color palettes to see their relationship with the reference values.
28 oct 2022
R-Ladies STL: Introduction to Quarto with Isabella Velásquez
27 oct 2022
22 oct 2022
11 oct 2022
SOIL SPECTROSCOPY GLOSOLAN WEBINAR
Interesting presentation from Leo-Ramirez-Lopez about the use of huge spectral libraries to select the proper samples to predict local samples using genetic algorithms that will be available on its R packages {rsamble} and {prospectr}., probably for the next year.
Github Tutorial
Managing - Part 2 (Github and RStudio) - RStudio
This is really a good tutorial about the use of RStudio and Github.
5 oct 2022
28 sept 2022
27 sept 2022
New Quarto blog for NIR and Chemometrics
I have created a new blog with Quarto, to work in R and R-Studio with spectroscopy
data available for Near Infrared . By the moment, a serial of posts are coming
about Near Infrared Transmitance with Meat data. It is a good ocasion to start
learning from the beginning to work with chemometrics in R with this type of
spectra
20 sept 2022
NIT Spectroscopic Tutorial with Caret (part 5)
Working these days with the tecator data set traying to find the better statistics as possible for the cross validation, and checking after with the test set. I use algorithms to remove the scatter but at the moment not derivatives to enhance the resolution. I show the statistics results for the CV and the validation results for the test set.
Multiple Scatter Correction has at the moment the better statistics for protein, but still some combinations of Math treatments could be tryed.
This is the XY plot for the test set with the pls model using the train set math treated with MSC.
16 sept 2022
NIT Spectroscopic Tutorial with Caret (part 4)
Looking to the spectra of the previous post was clear
that we must apply a pre-treatment to remove the baselines shifts. One way to
do it is to treat every spectrum individually, calculating a linear model
(absorbance values vs. wavelengths) and getting the values of the slope at
every wavelength, after these values are subtracted from the absorption values
of the spectrum getting the baseline corrected spectrum. We repeat the process
for every spectrum.
This function is available in the package pracma, and
its name is detrend. In case we use it for the spectra pre-process in Caret
with “center” and “scale”, we change the view from this:
To this:
This way we can see more clearly the variation in the
spectra that we can try to interpret some way. Also, with this new pre-treatment
we can check if the PCA change some way, and better to find outliers.
One of the options for interpretation are the
correlation spectrum (see the correlation of every wavelength with the
parameter of interest). In this case we can see how fat is inverse
correlated with protein and moisture, and there are some positive correlations
between protein and moisture.
We can see this if we run the correlation plot for the parameters;
12 sept 2022
NIT Spectroscopic Tutorial with Caret (part 3)
boxplot(train_scaled,
main = "preProcess(Center + Scale)")
In the plot we can see that are skewness to the right, and some outliers.
Now we
can apply the BoxCox algorithm together with Center and Scale and to look at
the boxplot spectra:
method = c("BoxCox", "center", "scale"))
boxplot(train_scaled_2,
The result shows how the skewness is removed. Of course, the PCA calculation will give different scores values, but still two terms will explain almost all the variance.
Looking to the spectra (treated either with “Center” and “Scale”, or “BoxCox”, “Center” and “Scale”) we see that there is a baseline shift that would be convenient that a pretreatment will remove in case we have to see clearly the variability of the fat, protein or moisture content.
6 sept 2022
NIT Spectroscopic Tutorial with Caret (part 2)
The first post of this tutorial brings a nice
conversation on tweeter. The idea now, is to use Caret with other packages (necessary for some of the prepocessing of the data), but I will work
in parallel at the same time with "tidymodels" packages (a good way
to learn how to use them).
Thanks to Max Kuhn for send me a link to some
interesting code from James Wade to use the Meat data in the tidyverse and
tidymodels environment. By the way the great book "Applied PredictiveModelling" (Max Kuhn & Kjell Johnson) is very useful and this tutorial
start as a possible solution for the exercise 6.1 in the book.
One of the problems to work with NIR or NIT spectra is the high collinearity of the predictors, this is due to NIR (Near Infrared Reflectance) and NIT (Near Infrared Transmitance) is formed by overtones (first, second, third) and combination fundamental bands which appear in the MIR (Middle Infrared). Other problem in this type of spectra is the high level of overlaping, and the NIR or NIT contains a lot of hidden information which needs from statistic or mathematical treatments to make them visible in a way that is usefull to develop models. This requires the use of preprocessing methods (centering, scaling, resolution improvement,...), that will be treated along the coming posts.
By the moment we will deal with the spectra without any treatment ("raw spectra"), and we create a correlation matrix to check the collinearity.
correlation <- cor(absorp)library(corrplot)
corrplot(correlation)
data_split <- createDataPartition(endpoints[ ,1], p = .75)
data_split <- data_split$Resample1
# split data
absorp_train <- absorp[data_split, ]
absorp_test <- absorp[-data_split, ]
- Center: The mean spectrum is sustracted from every spectrum.
- Scale : Every spectrum data point is divided by the standard deviation of all the data points in the spectrum.
We can see how center and scale affect to the spectra:
train_scaled <- scale(absorp_train, center = TRUE,
matplot(seq(850, 1048, by = 2), t(train_scaled),
xlab = "Wavelengths", ylab = "Absorbance",
main = "Meat spectra", type = "l")
These pretreatments are aplyed to every spectrum individually, so at the end of the preprocess (before developing the PCA) we have a transformed data matrix with the same dimensions (215 . 100).
pca_object <- prcomp(absorp_train, center = TRUE,
percent_variance <- pca_object$sdev^2/sum(pca_object$sd^2)*100
plot(percent_variance[1:20])
head(percent_variance)
head(pca_object$x[1:5 , 1:2 ]) #scores with just 2 terms
wavelengths <- seq(850, 1048, by = 2)
colnames(absorp_train) <- wavelengths
colnames(absorp_train)
trans <- preProcess(absorp_train,
trans #info about the PCA calculation
transformed <- predict(trans, absorp_train)
head(transformed[1:2 , ])
5 sept 2022
NIT Spectroscopic Tutorial with Caret (part 1)
Along several posts we will practice the use of Caret with spectroscopic data from an Infratec Instrument. The first thing we will do, is to load the library and the data which contain the spectroscopic and reference values.
#Loading the datadata(tecator)
?caret::tecator #Details about the tecator data
Two tables are loaded, one with the spectroscopic data (absorp), and another with the reference values (endpoints).
dim(endpoints)dim(absorp)
We can see that we have 215 samples measured at 100 data points in the spectroscopic data, and three parameters with reference values.
The data points goes from 850 to 1048 every two nm, so they are in total 100 data points, and the reference values are for Moisture, Fat and Protein. As we can read in the Help info, the samples are chopped pure meat measured in transmittance.
We can see first the structure of the data is numeric
str(absorp)str(endpoints)
and their class (matrix)
class(absorp)class(endpoints)
We have a matrix for the absorption values and a matrix for the reference values. Let´s see the raw spectra
matplot(seq(850, 1048, by = 2), t(absorp),
xlab = "Wavelengths", ylab = "Absorbance",
main = "Meat spectra", type = "l")
As we can see, due to the particle size, differences in the pathlength and other physical circumstances, there is quite a lot scatter in the spectra, so this physical variables are with others (chemical) data that we will have to treat and manage to develop models.
13 ago 2022
Soviet Space Dogs
These dogs were found in the street and trained to go to the space in really very hard conditions.
I use the template to prepare this graph where we see their ocupants (normally two dogs) and how in the sixtys several trips had succedd going into orbit for several days.
There are 5 flights not included due that the altitude of the flight is unknown.
10 ago 2022
6 ago 2022
#tidytuesday:Eurovision (2022 Final Televote Votes)
See the group itself one for Ukraine, and see the case for Serbia.
#tidytuesday:Eurovision (2022 Final Jury Votes)
There is some polemic with the votes in Eurovision Final 2022 with some agreements possibly within some countries. Data Science can help us to see patterns within the countries, checking the clusters and with that base investigate more.
I show
the cluster dendogram for the Final Votes from the Jury this year 2022.
5 ago 2022
4 ago 2022
#tidytuesday:Eurovision (Spanish Artist and their positions)
As we see in the plot Conchita Velasco and Raphael repeat representing Spain on Eurovision. The colour in the plots show their position (greener the better).
spanish_artists <-
eurovision_1 %>%
filter(section=="final"| section=="grand-final") %>%
filter(artist_country == "Spain") %>%
select(year, artist, song, host_city, host_country, rank)
spanish_artists %>%
group_by(year) %>%
mutate(artist = fct_reorder(artist, year)) %>%
ggplot(aes(x = year, y = artist)) +
geom_tile(aes(fill = rank), colour = "red") +
scale_fill_gradient(low="green", high="red", limits=c(1, 26)) +
scale_x_continuous(breaks = seq(1956, 2022)) +
theme(axis.text.x = element_text
(angle = 90, vjust = 0.5, hjust = 1)) +
labs(y = "Spanish artist on Eurovision by year", y = "Year")
3 ago 2022
#tidytuesday:Eurovision (What happened in 1956?)
Another special year. It was the first edition of Eurovision Festival with seven participant countries (founders) and two artists representing each country with different songs, except Switzerland and Luxemburg with just one singer for the two songs.
Lys Assia from Switzerland was the winner.
There is a nice explanation on Wikipedia about that edition.
eurovision_1 %>%filter(year == 1956) %>%
count(artist_country, sort = TRUE)
A tibble: 7 × 2
artist_country n
<chr> <int>
Belgium 2
France 2
Germany 2
Italy 2
Luxembourg 2
Netherlands 2
Switzerland 2
All this must be taken into account when working with some statistics with the Eurovision data base.
2 ago 2022
#tidytuesday:Eurovision (What happened in 1969?)
It was a special year with four winners:
eurovision_1 %>%filter(section=="final"| section=="grand-final", winner == TRUE) %>%
count(winner, year, host_country, sort = TRUE)
50 years ago today: Four winners at Eurovision 1969 in Madrid
#tidytuesday: Eurovision
Another question we can ask about the history of Eurovision Festival is: which was the country which host the festival more often?
Normally the host country is the one which won the previous year, but for different reasons some years is not like that. That will be the case for 2023 where the host country will be United Kingdom (2nd place in 2022) indeed Ukraine for obvious reasons (1st place in 2022).
As we can see in the plot UK is the country which host more ofen the Eurovision Festival.
countries_events <-unique(eurovision_1 %>%
group_by(host_country) %>%
summarize(host_country, event, year))
countries_events %>%
count(host_country, sort = TRUE) %>%
ggplot(aes(x = reorder(host_country, n), n)) +
geom_col() +
coord_flip()
#tidytuesday: Eurovision
From today there is a new label in the blog to work with the tidytuesday data available from tidytuesday.
There are a lot of tutorials to learn to tidy our data to prepare it to extract information, and to develop models (I normally see the videos of Julia Silge and David Robinson), so even if these post are not much related to the world of chemometrics they will help us to have fun with the "R" language, get better skills and habits that we will use it when necessary to work with R in our daily work.
I select the Eurovision data in this case, and the first thing I ask myself was to try to find a plot which shows me the evolution of the Eurovision contest
eurovision_1 %>%count(year,section, sort = TRUE) %>%
ggplot(aes(year, n, fill = section )) +
geom_col(width = 1, colour = "black") +
theme(axis.text.x = element_text(angle = 90, vjust = 0.1)) +
scale_x_continuous(breaks = seq(1956, 2022)) +
theme(axis.text=element_text(size=7)) +
coord_flip()
28 jul 2022
Convert Nanometers to Wavenumbers
All we know that the wavelength scale (X axis) is different for a dispersive NIR or a FTNIR. Sometimes, for some reasons we want to convert the wavelength scale of a NIR spectra database, to the wavenumber scale or vice versa.
In this case I apply with R the conversion of the NIRsoil$spc data (from the "prospectr" package) from nanometers :
We just have to apply the formula:
12 jul 2022
NIR RMS between subsamples
One of the advantages of R, is that you can develop a function to apply to your calculations to get your purpose. In this case I wanted to apply the RMS calculation to every row of the difference matrix we have seen in the previous post.
We can see with an histogram the distribution of the RMS values
11 jul 2022
Diference between subsamples
Once we have sorted and grouped all the samples and subsamples, we can create two matrices one that contains the odd samples (first subsample) and other with the even subsamples (second subsample), this way we can calculate the spectra difference between subsamples and check the spectra.
This difference
spectra matrix is important to calculate the RMS between subsamples, that gave
us an idea how similar are the subsamples between them. Just sum the squared values
for every spectrum difference, divide by the number of wavelengths and
calculate the square root.
8 jul 2022
6 jul 2022
Soil clay regressions: Looking for the better accuracy (part 3)
Now we have four calibrations for clay in soil developed with a selection of samples from the LUCAS database (Spanish crop soils). Now we want to see if those calibrations predict with certain accuracy a new set of soil samples (155) from a Spanish region, acquired in a different instrument and at a different laboratory.
In these cases, it is normal to expect a bias or a slope
in the predictions, so we can use the model with those adjustments applied, until
the database is updated and a new expanded method with new variability
(instrument, laboratory, region,) developed.
Well, these are the results of the XY plots "Lab vs
Predictions" for this independent data set:
PLS predictions:
Cubist Predictions
MBL Predictions
As we see in all the cases some adjustment or
calibration update is needed. We can try to reprocess everything trying to find
a better configuration which improves these values, but in that case this new
set will never be independent again like it is now.
5 jul 2022
Soil clay regressions: Looking for the better accuracy (part 2)
Let´s select a
seed (to fix the training and test set) and develop the regression with four
algorithms (PLS, Random Forest, Cubist and Memory Based Learning.
These are the
Test Validation XY plots and statistics:
PLS Regression:
Cubist Regression:
Memory Based Learning Regression
We can see PLS give the better RMSEP, but some samples are outside the Action Limits Warning, while that in the MBL the residual distribution is more stable and there are no samples outside the action limits threshold.
Questions about NIR Modelling (001)
Sometimes I
receive mails from the readers, that are very interesting, so I create this
post to answer the reader and to keep the post to create comments or add what
the readers consider about their own experience.
The choice of the wavelength corresponding to the studied parameter (is it better to keep all the scan or to choose a part which represents the targeted parameter? If any, how to do so?)
Normally all the scan is used, and the PLS algorithm, latent
variables (PLS terms) which represents the spectral variance and their covariance
with the studied parameters. Looking to the regression coefficients, and
knowing the wavelengths at which those correlation absorb, you can try to
interpret the regression and decide if certain wavelength zones could be
excludes (as flat zero zones, …..). Regression coefficients are very difficult
to interpret due to the math treatments applied (specially derivatives).
Other option is to choose a few specific wavelengths,
when normally the first one is the one at which the parameter absorbs (example:
1940 nm for the water) and continue adding wavelengths of other constituents
that interfere with the water, or zones that do not absorb, but scatter is
observed. Normally the software helps you with these selections, and you have always
the statistics to see if the wavelengths added improve the regression. Normally
this type of algorithm is called MLR (Multiple Linear Regression).
How to split the samples between calibration and validation (is there a test to do?)
Split randomly 80% of the samples to the Training Set,
and the remain 20% to the Test Set. In the case you have a lot of samples from
different years you can uses other approaches (the older samples for
calibration and the new ones for validation, …..). Anyway, if the calibration
is robust, you should get similar results.
The criteria for choosing the tests to be performed for pre-processing (2nd derivative, SNV, MSC, etc.).
Normally the criteria is to choose the simplest math
treatment. For the scatter, if you have a lot od samples and all the possible
variability represented you can try MSC, if not one of the best options is to
combine SNV and Detrend.
First derivative is difficult to interpret (the maximum
for the raw spectra, becomes a zero crossing), second derivative is better
interpretable. The important option is the gap you use (not to long because you
can loose information, and not too short because you add noise).