In the case of the loadings for PLS we do not have the condition of orthogonality as in the weights or scores, even if they are coming from orthogonal scores.
So the product of the matrices Pt.P
is not a diagonal matrix, and this condition is what makes PLS regression special and different from Principal Components Regression.
Let´s check:
Xodd_pls3_ld1
Xodd_pls3_ld2
Xodd_pls3_ld3
Xodd_pls3_ld4
p.matrix<-cbind(Xodd_pls3_ld1,Xodd_pls3_ld2,
Xodd_pls3_ld3,Xodd_pls3_ld4)
> round(t(p.matrix) %*% p.matrix,4)
Xodd_pls3_ld1 Xodd_pls3_ld2 Xodd_pls3_ld3 Xodd_pls3_ld4
Xodd_pls3_ld1 1.0132 -0.1151 0.0000 0.0000
Xodd_pls3_ld2 -0.1151 1.2019 -0.4493 0.0000
Xodd_pls3_ld3 0.0000 -0.4493 1.8783 -0.9372
Xodd_pls3_ld4 0.0000 0.0000 -0.9372 1.0134
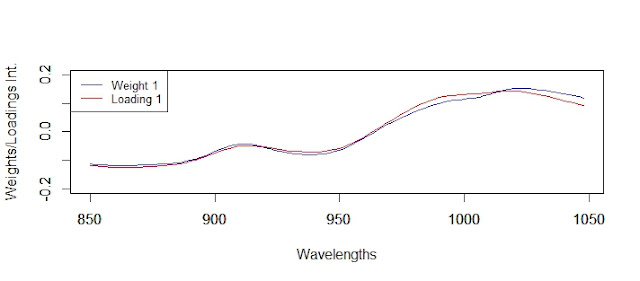
We can see the red numbers that makes that we don´t get the condition of orthogonality, and how the loadings 2 and 3 are specially involved, and these is because in this case the loadings 1 and 4 are very similar to the weights 1 and 4, and 2 and 3 are quite different.
So loadings 1 and 4 are orthogonal between them. So there is no condition for the loadings.
Xodd_pls3_ld1 Xodd_pls3_ld2 Xodd_pls3_ld3 Xodd_pls3_ld4
Xodd_pls3_ld1 1.0132 -0.1151 0.0000 0.0000
Xodd_pls3_ld2 -0.1151 1.2019 -0.4493 0.0000
Xodd_pls3_ld3 0.0000 -0.4493 1.8783 -0.9372
Xodd_pls3_ld4 0.0000 0.0000 -0.9372 1.0134
We can see the red numbers that makes that we don´t get the condition of orthogonality, and how the loadings 2 and 3 are specially involved, and these is because in this case the loadings 1 and 4 are very similar to the weights 1 and 4, and 2 and 3 are quite different.
So loadings 1 and 4 are orthogonal between them. So there is no condition for the loadings.