Hi all, I am quite busy so I have few time to
expend on the blog, anyway I have continue working with R trying to develop
functions in order to check if our models perform as expected or not.
Residual plots and the limits (UAL,UWL,LWL,UAL) we draw
on them will help us to take decisions, but developing some functions can help
us to see suggestions in order to take good decisions.
So I am trying to works on this.
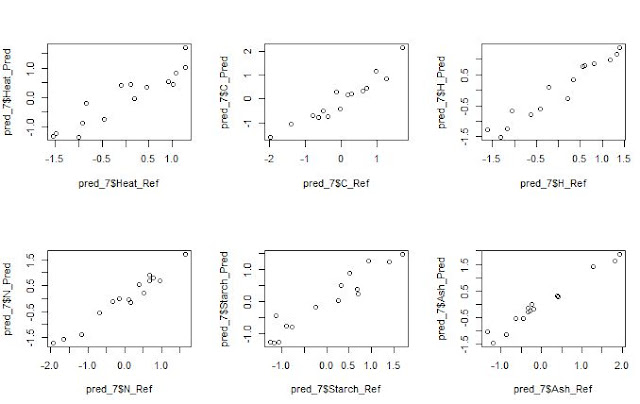
We always want to compare the results from a Host to a
Master, the predicted NIR results with the Lab results,….
In all these predictions we have to provide realistic
statistics and not too optimistic, if not we will not understand really how our
model performs. Validation statistics, and looking to the residual plots
will help us to understand if: our standardization is performing fine, if we
have a bias problem or if the samples of the validation should be include in
the data set and recalibrate again.
In this case is important to know the RMSEP of our calibration
which can be the SECV for example (standard error of cross validation), and
compare this error with the RMSEP of the validation, and after this with the
SEP (validation error corrected by bias).
Is important to see how the samples are distributed in the residual plot into
the warning limits (UWL and LWL) and into the action limits (UAL and LAL), are
they distributes randomly?, do they have a bias?, if I correct the bias the
distribution becomes random and into limits?,.....There are several questions
that if we have the correct answer will help us to improve the model, and to
understand and explain to others the results we obtain.
This is a case where the model performs with a Bias:
Validation
Samples = 9
RMSEP : 0.62
Bias : -0.593
SEP : 0.189
Corr : 0.991
RSQ : 0.983
Slope : 0.928
Intercept:
0.111
RER : 18.8
Fair
RPD : 7.02
Excellent
BCL(+/-)
: 0.143
***Bias
adjustment is recommended***
The residual plot confirms that we have a bias:
Using
SEP as std dev the residual distibution is:
Residuals into 68% prob (+/- 1SEP) = 0
Residuals into 95% prob (+/- 2SEP) = 1
Residuals into 99.5% prob (+/- 3SEP) = 4
Residuals outside 99.5% prob (+/- 3SEP) = 5
Samples outside UAL = 0
Samples outside UWL = 0
Samples inside WL =
1
Samples outside LWL = 8
Samples outside LAL = 5
With
Bias correction the Residual Distribution would be:
Residuals into 68% prob (+/- 1SEP) =7
Residuals into 95% prob (+/- 2SEP) =9
Residuals into 99.5% prob (+/- 3SEP) =9
Residuals out
99.5% prob (> 3SEP) =0
With the bias correction
the statistics are better and confirm that probably a non robust
standardization has been done with these two instruments that we are comparing.
This can help us to check
other standardizations or decide if we need other algorithms as repeatibility
file in the calibration or to mix spectra from both instruments.