We can coloured the scores by their season value, so we can see how every season extend the scores in the different principal components maps.
fish_1d_all_pc<-nipals(fish_1d_all$nir_1d,10)
rownames(fish_1d_all_pc$T)<-fish_1d_all$Sample
colnames(fish_1d_all_pc$T)<-seq(1,10,by=1)
##### Creating a list for NIPALS
library(chemometrics)
fish12_nipals <-list(scores=fish_1d_all_pc$T,
loadings=fish_1d_all_pc$P,
sdev=apply(fish_1d_all_pc$T, 2, sd))
Now we plot the orthogonal and scores distances and we check which ones are outside certain limits, coulouring the samples by their season and using 5 principal components.
res<-pcaDiagplot(fish_1d_all$nir_1d,X.pca=fish12_nipals,
a=5,col=fish_1d_all$Season)
Samples outside the cutoff are the Mahalanobis distance outliers considering 5 principal components.
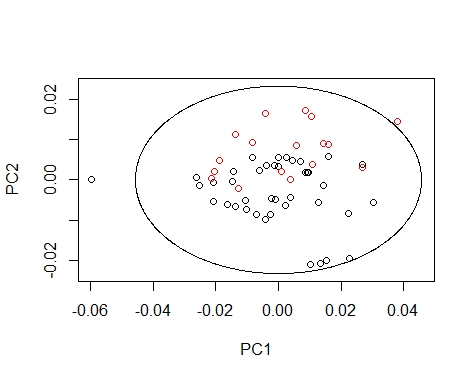
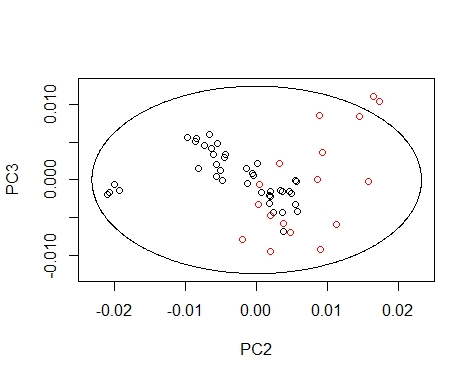
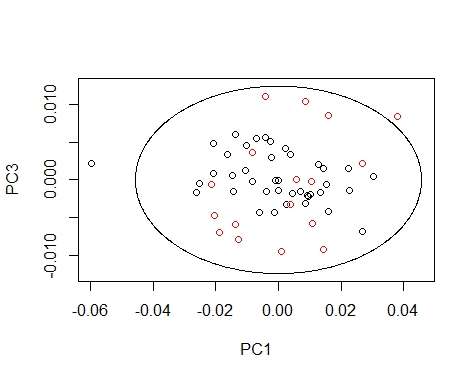
We can draw the Mahalanobis Ellipses with the combinations: PC1 vs PC2,
PC1 vs PC3, and PC2 vs PC3. Samples are coloured by their season (black for fish1 and red for fish2).
library(chemometrics)
par(mfrow=c(1,1))####### PC2 vs PC3 ##########
drawMahal(fish_1d_all_pc$T[,c(1,2)],
center=apply(fish_1d_all_pc$T[,c(1,2)],2,mean),
covariance=cov(fish_1d_all_pc$T[,c(1,2)]),
quantile=0.975,xlab="PC1",ylab="PC2",
####### PC2 vs PC3 ##########
drawMahal(fish_1d_all_pc$T[,c(2,3)],
center=apply(fish_1d_all_pc$T[,c(2,3)],2,mean),
covariance=cov(fish_1d_all_pc$T[,c(2,3)]),
quantile=0.975,xlab="PC2",ylab="PC3",
col=fish_1d_all$Season)
####### PC1 vs PC3 ########
drawMahal(fish_1d_all_pc$T[,c(1,3)],
center=apply(fish_1d_all_pc$T[,c(1,3)],2,mean),
covariance=cov(fish_1d_all_pc$T[,c(1,3)]),
quantile=0.975,xlab="PC1",ylab="PC3",
col=fish_1d_all$Season)
Samples out of the ellipse can be identified with the identify function but that does not mean that it is an outlier because that depends of a general computation between all the principal components.
As you can see all the process is becoming very straightforward as we continue with more posts of more fishing seasons.

No hay comentarios:
Publicar un comentario