In the case of the Soy meal, we have a validation sample set (from an Infratec Nova), and we get the predictions from a calibration sample set from an Infratec 1241. As we saw in "Analyzing Soy meal in transmittance (part 8)" , the predictions are fine, but we have a bias and the idea was to merge the samples from the Infratec Nova with the samples from the Infratec 1241 and to develop a calibration.
But before that we want to see another option, and for that we are going to use an option from Win ISI: "Select local samples from a product file".
With this option we project the validation samples in the PC space we got from the calibration samples, and we search for neighbors into a certain cutoff.
Those samples will be take them apart into a file, to develop an exclusive calibration for the validation samples.
In this case I am going to use a cutoff of 0,2 (Mahalanobis distance).
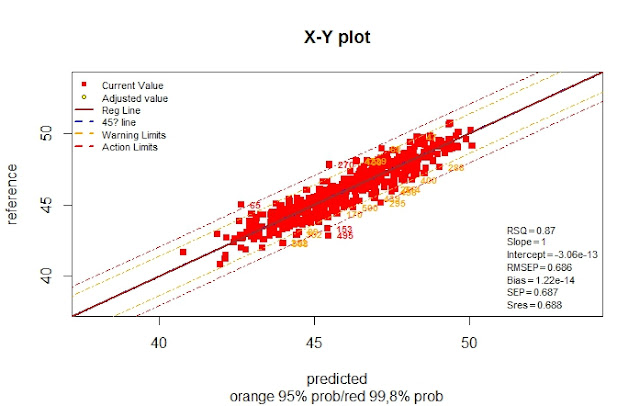
From the 657 samples, 527 were selected and exported to R to develop the calibration. There are some clear outliers, but there is a high improve in the statistics.
Now we remove the samples with number in red, because they are out of the action limits and recalculate.
>soy_ift_prot_sel1_r1<-soy_ift_prot_sel1[-c(495,102,83,270,74),]
>Prot_plsr_sel_r1<-plsr(soy_ift_prot_sel1$Prot~soy_ift_prot_sel1 $Xsel_msc,ncomp=16,data=soy_ift_prot_sel1,validation = "LOO")
>predictions_sel_r1<-(Prot_plsr_sel_r1$fitted.values[,,9])
>soy_ift_prot_sel2_r1<-cbind(soy_ift_prot_sel1_r1$Sample,soy_ift_prot_sel1_r1$Prot,predictions_sel_r1)
>soy_ift_prot_sel2_r1<-cbind(soy_ift_prot_sel1_r1$Sample,soy_ift_prot_sel1_r1$Prot,predictions_sel_r1)
>monitor_prot_sel<-monitor10c24xyplot(soy_ift_prot_sel2_r1)
As you can see there is an improvement in the calibration statistics with this selection, but is it an improvement in the validation. We will see it in the next post.

No hay comentarios:
Publicar un comentario