I have ten new sunflower seed samples, with laboratory data and I´m going to use them to validate the performance of a model developed in R with PLS:
Sunflower seed Regressions with "R" - 001
Sunflower seed Regressions with "R" - 001
First, I have a look to the spectra of the validation set (red spectra) compares with the training spectra (blue spectra), without any math treatment applied:
and after, with the MSC applied:
I see clearly some differences, but the idea is to check if the calibration is robust enough to predict the samples according to the statistics we got in the summary of the regression.
In the summary of Sunflower seed Regressions with "R" - 001 , we decide to use 7 terms for our predictions, so:
predict(sflw.g00rmn,ncomp=4,newdata=sflw.msc2.val)
G00rmn
171 46.25923
173 53.07202
176 53.48508
177 53.27027
178 46.05511
179 46.73826
180 50.95862
181 52.44956
182 47.59493
183 46.51557
The error is:
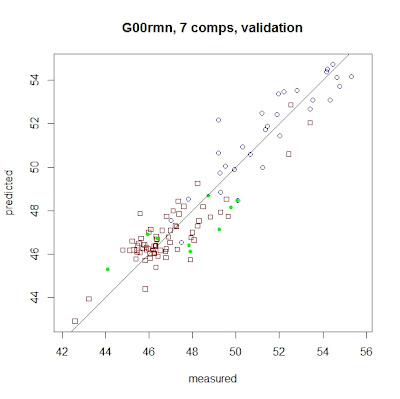
Let´s have a look to the "Reference vs Predicted" plot:
predplot(sflw.g00rmn,ncomp=7,newdata=sflw.msc3.val,
asp=1,line=TRUE,col=c("red"))
and after, with the MSC applied:
I see clearly some differences, but the idea is to check if the calibration is robust enough to predict the samples according to the statistics we got in the summary of the regression.
In the summary of Sunflower seed Regressions with "R" - 001 , we decide to use 7 terms for our predictions, so:
predict(sflw.g00rmn,ncomp=4,newdata=sflw.msc2.val)
G00rmn
171 46.25923
173 53.07202
176 53.48508
177 53.27027
178 46.05511
179 46.73826
180 50.95862
181 52.44956
182 47.59493
183 46.51557
The error is:
Let´s have a look to the "Reference vs Predicted" plot:
predplot(sflw.g00rmn,ncomp=7,newdata=sflw.msc3.val,
asp=1,line=TRUE,col=c("red"))