In the Caret package, we have a data set called “tecator”
with data from an Infratec for meat. In the book “Applied Predictive Modelling”,
is used as an exercise in the Chapter : “Linear Regression and its Cousins”, so I´m
going to use it in this and some coming
posts.
When we develop a PLS equation with the function “plsr”
in the “pls” package we get several values, and one of them is “validation”,
where we get a list with the predictions for the number of terms selected for
the samples in the training set. With these values, several calculations will
define the best model, so we does not overfit it.
Anyway, to keep apart a random set for validation will
help us to adopt the best decision for the selection of terms. For this, we can
use the “createDataPartition” from the Caret package. The “predict” using the
developed model and the external validation set will give us the predictions
for the external validation set and comparing this values with the reference
values we will obtain the RMSE (using
the RMSE) function, so we can decide the number of terms to use for the model
we use finally in routine.
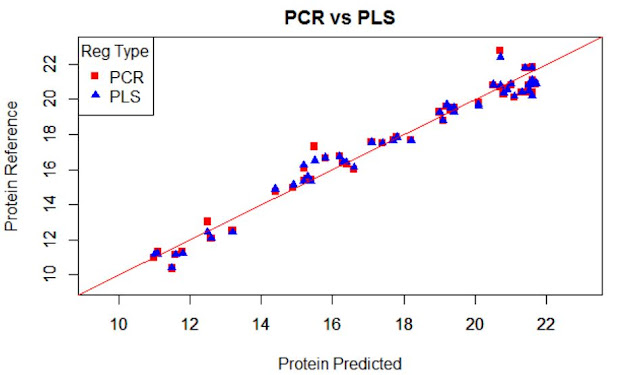
We normally prefer plots to see performance of a
model, but the statistics (numbers) will really decide how the performance is.
In the case I use 5 terms (seems to be the best option),
the predictions for the training set are:
train.dt.5pred<-plsFitdt.moi$validation$pred[,,5]
In addition, the predictions for the external
validation set are:
test.dt.pred<-predict(plsFitdt.moi,ncomp=5,
newdata = test.dt$NIR.dt)
With these values
we can plot the performance of the model:
plot(test.dt.pred,test.dt$Moisture,col="green",
xlim=range.moi,ylim=range.moi,
ylab="Reference",xlab="Predicted")
par(new=TRUE)
plot(train.dt.5pred,tec.data.dt$Moisture,col="blue",
xlim=range.moi,ylim=range.moi,
xlab="",ylab="")
abline(0,1,col="red")
Due to the high range of this parameters we can see plots as this, where we can see area ranges with bias, others with more random noise, or others with outliers.