This is a post to follow the example of the cereal data
in the book “Introduction to multivariate Statistical Analysis in Chemometrics”
(Kurt Varmuza & Peter Filzmoser).
PLS2 is applied to a Cereal Data set with the function “mvr”
using the cross validation “LOO: leave one out”.
The validation plot shows the error of the validation vs the
number o PLS terms, for the different constituents.
A for every constituent the number of terms choose can be
different, so an average is recommended.
This plot shows the error of the validation vs the number
o PLS terms, for all the constituents overlapped.
As we can see, we choose 7 components for the model.
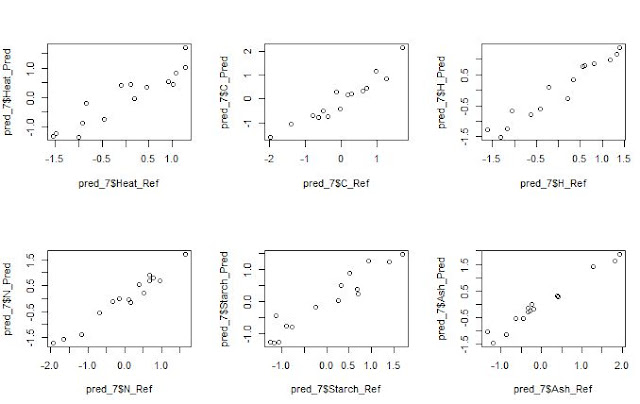
We can see the XY plots (reference vs predicted) with 7
terms for all the constituents.
pred7<-predict(respls2,
cereal, ncomp = 7,type = c("response"))
par(mfrow=c(2,3))
plot(pred_7$Heat_Ref,pred_7$Heat_Pred)
plot(pred_7$C_Ref,pred_7$C_Pred)
plot(pred_7$H_Ref,pred_7$H_Pred)
plot(pred_7$N_Ref,pred_7$N_Pred)
plot(pred_7$Starch_Ref,pred_7$Starch_Pred)
plot(pred_7$Ash_Ref,pred_7$Ash_Pred)